30th International European Conference on Parallel and Distributed Computing (Euro-Par 2024)
DOI: TBC
PDF Version
Abstract
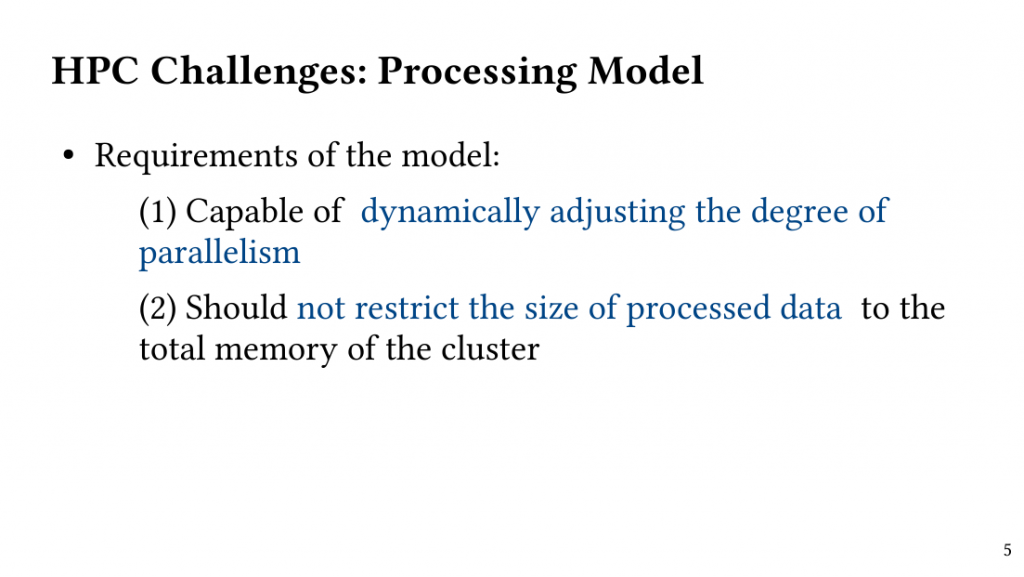
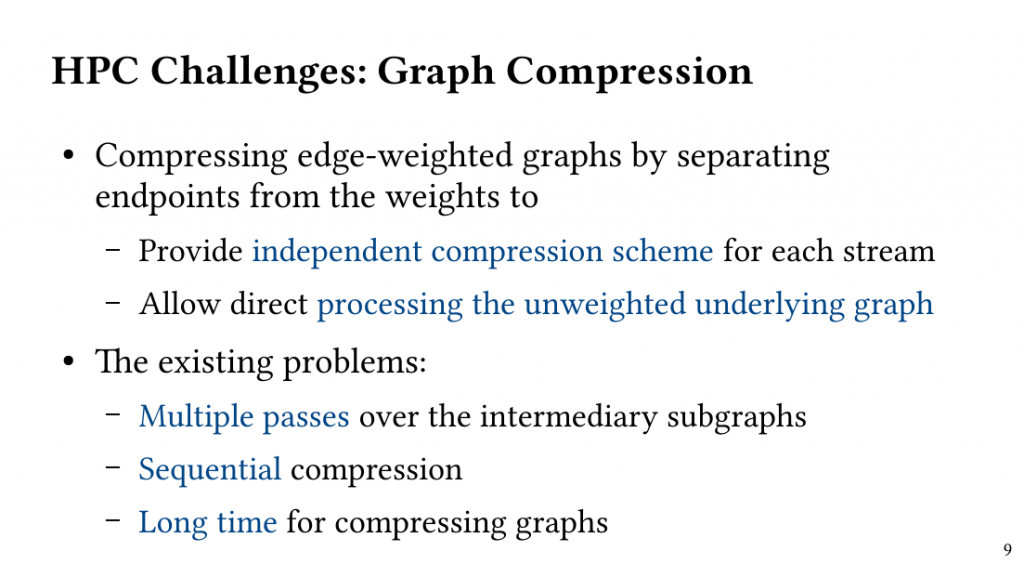
The Maximum Weighted Clique(MWC) problem remains challenging due to its unfavourable time complexity.In this paper, we analyze the execution of exact search-based MWC algorithms and show that high-accuracy weighted cliques can be discovered in the early stages of the execution if searching the combinatorial space is performed systematically.
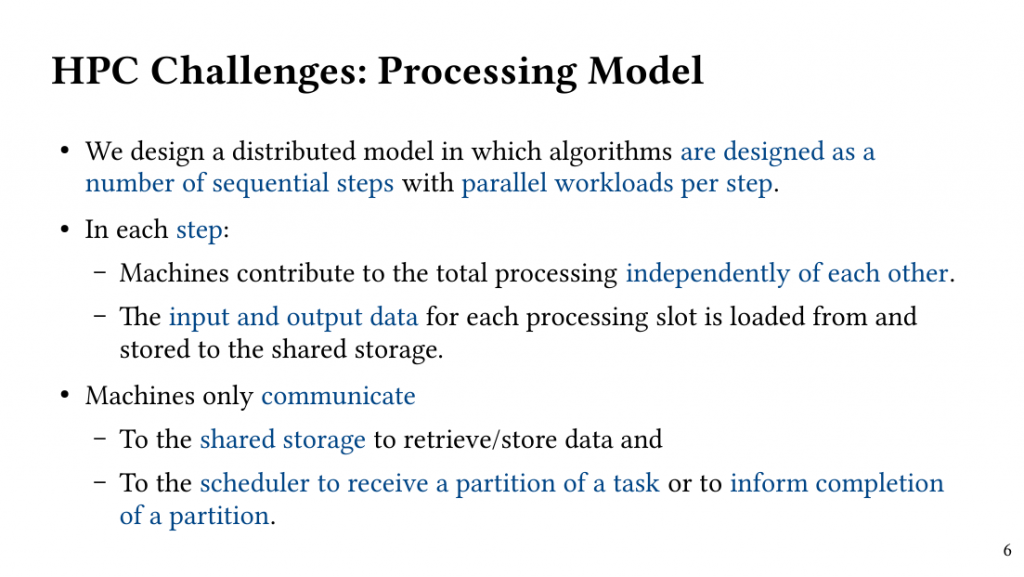
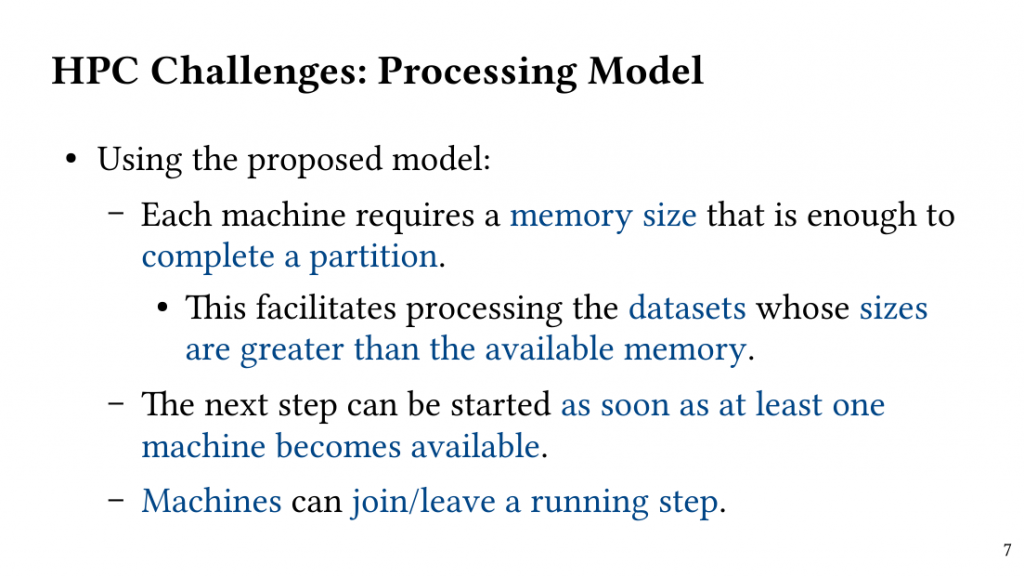
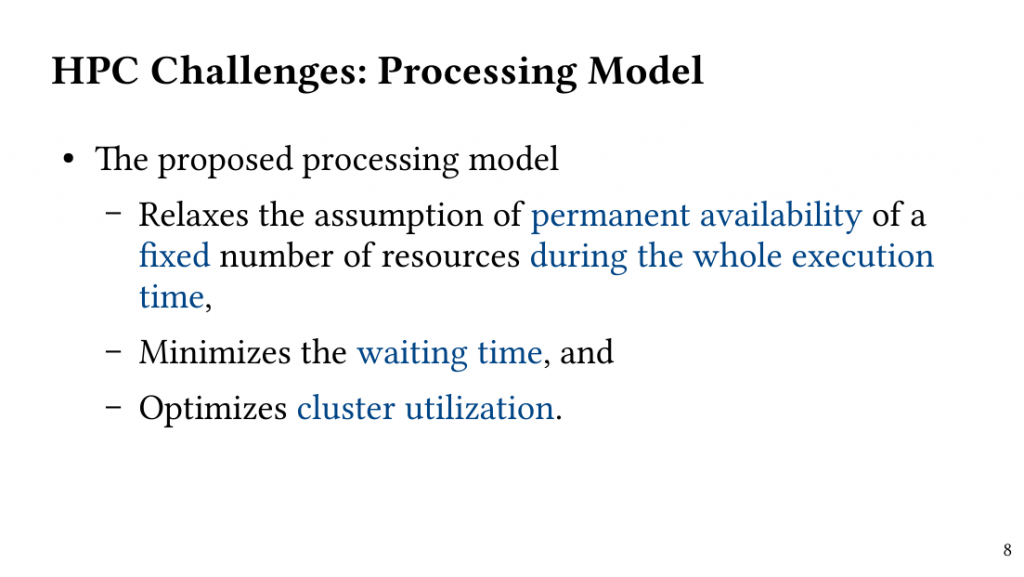
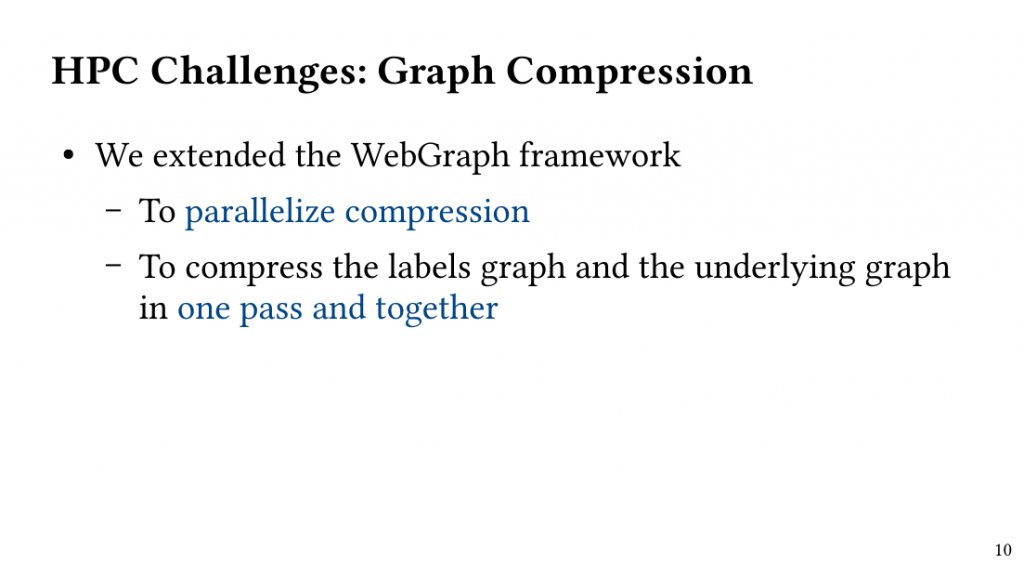
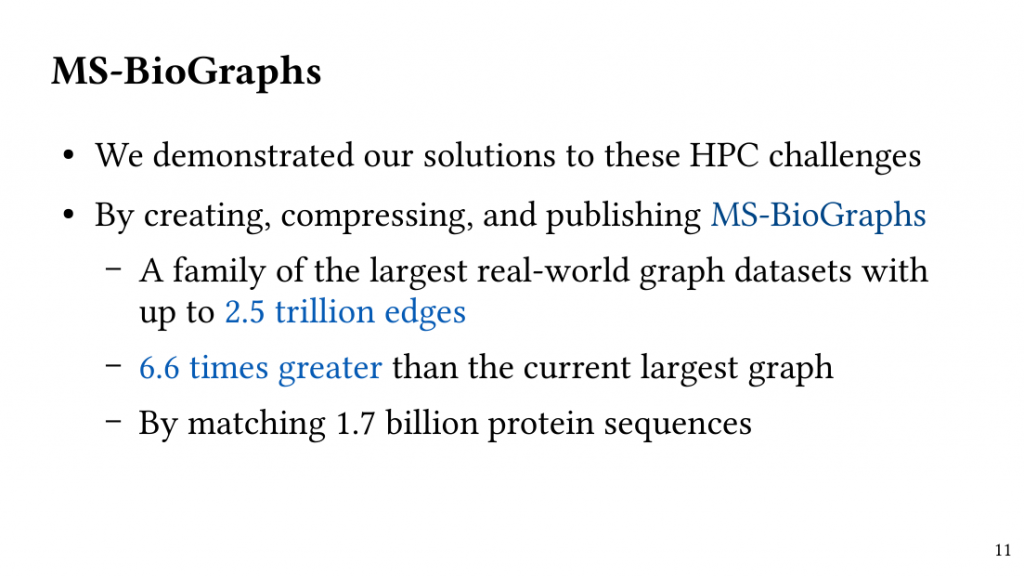
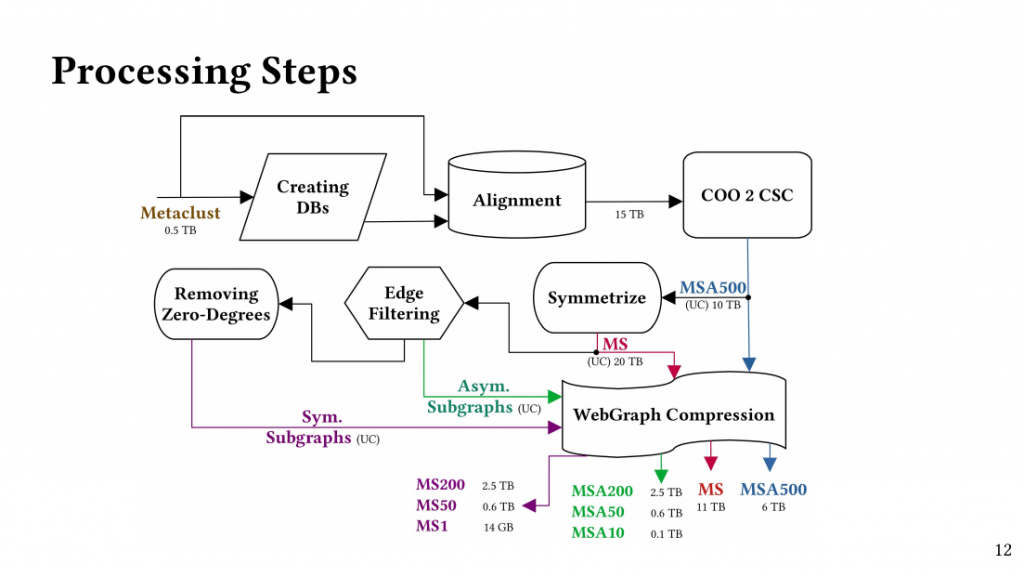
Based on this observation, we introduce QClique as an approximate MWC algorithm that processes the search space as long as better cliques are expected. QClique uses a tunable parameter to trade-off between accuracy vs. execution time and delivers 4.7-$82.3 time speedup in comparison to previous state-of-the-art MWC algorithms while providing 91.4% accuracy and achieves a parallel speedup of up to 56x on 128 threads.
Additionally, QClique accelerates the exact MWC computation by replacing the initial clique of the exact algorithm. For WLMC, an exact state-of-the-art MWC algorithm, this results in 3.3x on average.
Code