DOI:
Whereas the literature describes an increasing number of graph algorithms, loading graphs remains a time-consuming component of the end-to-end execution time. Graph frameworks often rely on custom graph storage formats, that are not optimized for efficient loading of large-scale graph datasets. Furthermore, graph loading is often not optimized as it is time-consuming to implement.
This shows a demand for high-performance libraries capable of efficiently loading graphs to (i) accelerate designing new graph algorithms, (ii) to evaluate the contributions across a wide range of graph datasets, and (iii) to facilitate easy and fast comparisons across different graph frameworks.
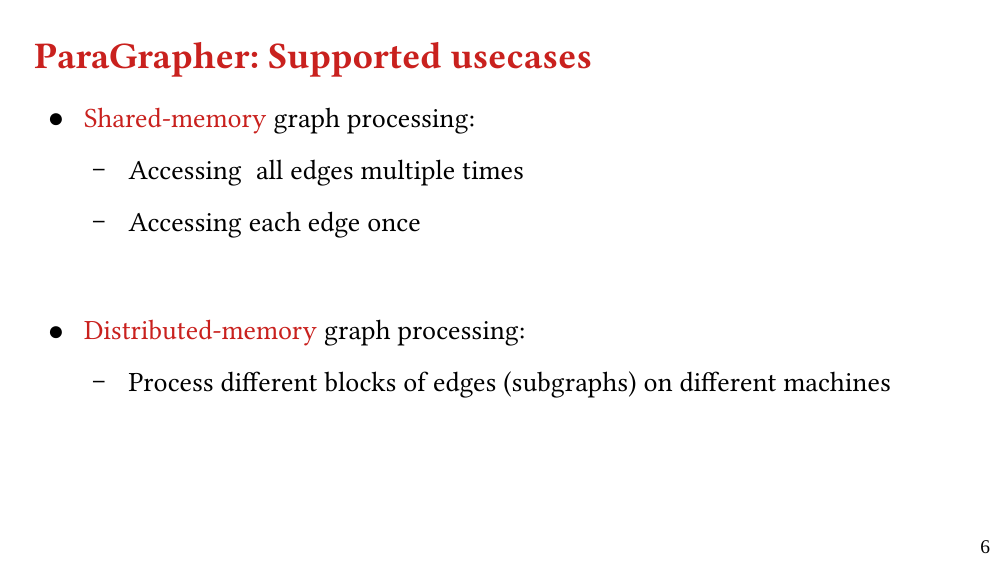
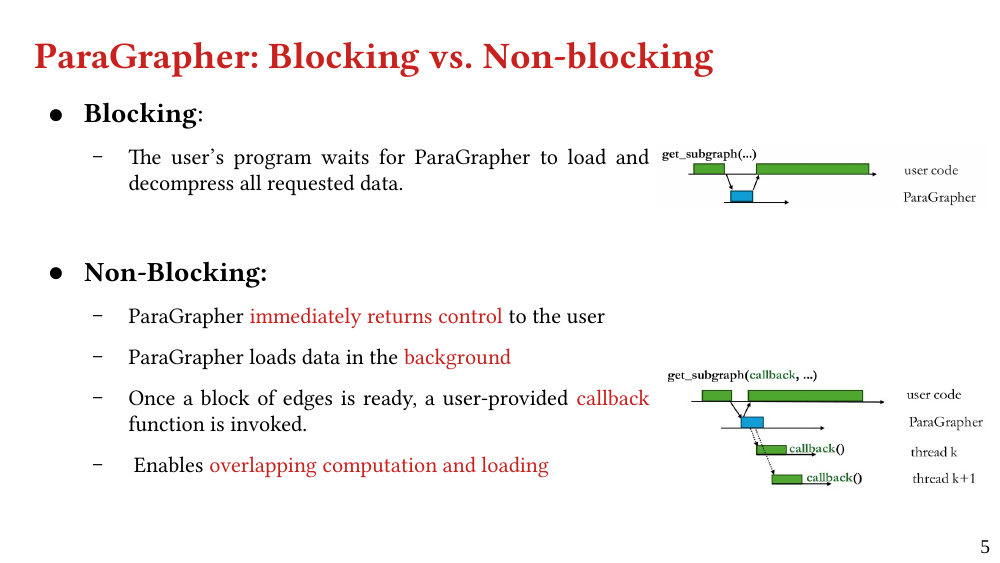
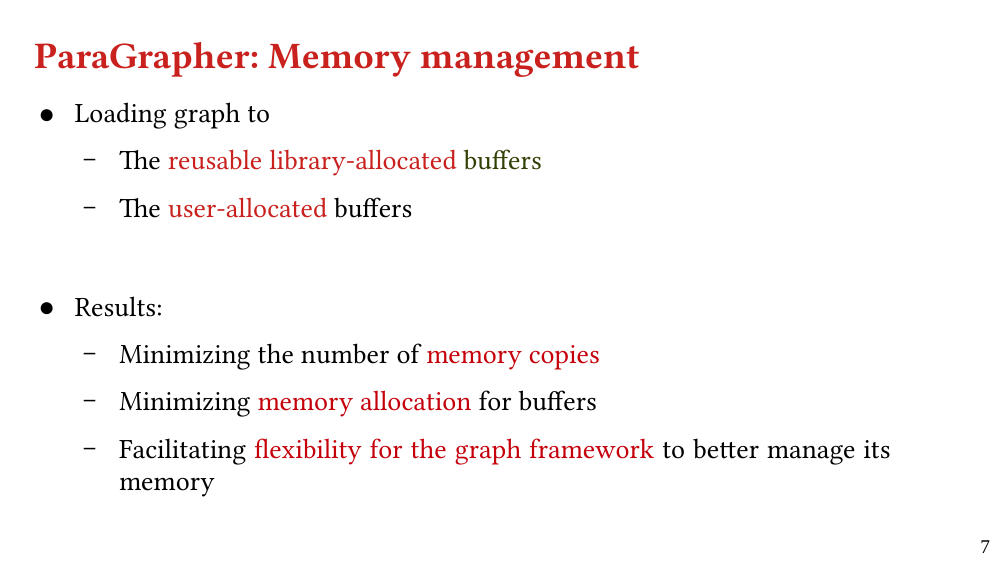
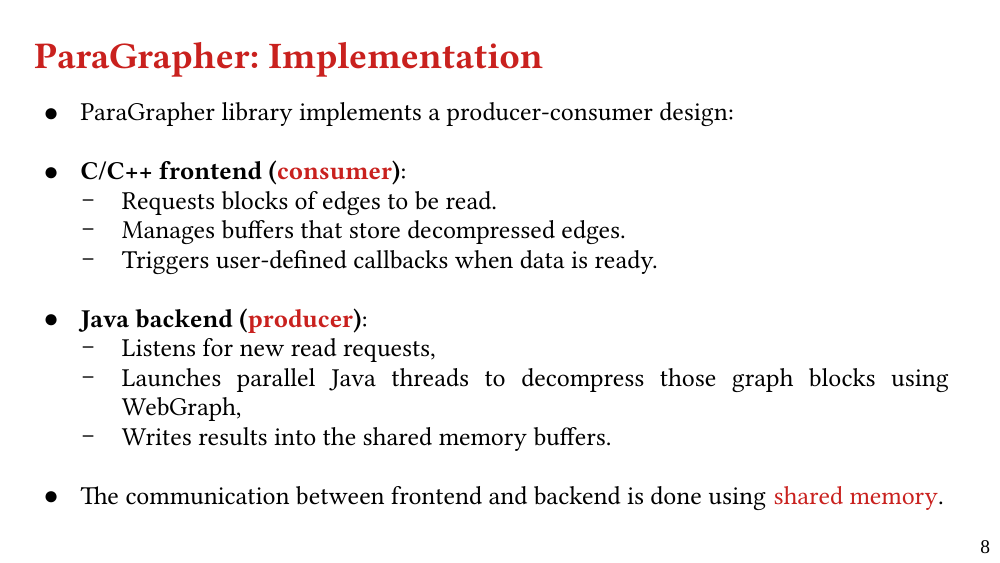
We present ParaGrapher, a library for loading large-scale compressed graphs in parallel and distributed graph frameworks. ParaGrapher supports (a) loading the graph while the caller is blocked and (b) interleaving graph loading with graph processing. ParaGrapher is designed to support loading graphs in shared-memory, distributed-memory, and out-of-core graph processing.
We explain the design of ParaGrapher and present a performance model of graph decompression. Our evaluation shows that ParaGrapher delivers up to 3.2 times speedup in loading and up to 5.2 times speedup in end-to-end execution (i.e., through interleaved loading and execution).
Source Code
https://github.com/DIPSA-QUB/ParaGrapher
API Documentation
Please refer to the Wiki, https://github.com/DIPSA-QUB/ParaGrapher/wiki/API-Documentation, or download the PDF file using https://github.com/DIPSA-QUB/ParaGrapher/raw/main/doc/api.pdf .
BibTex
@articel{paragrapher-bigdata,
}Related Posts & Source Code
- ParaGrapher: A Parallel and Distributed Graph Loading Library for Large-Scale Compressed Graphs – BigData’25 (Short Paper)

- Accelerating Loading WebGraphs in ParaGrapher

- Selective Parallel Loading of Large-Scale Compressed Graphs with ParaGrapher – arXiv Version

- An Evaluation of Bandwidth of Different Storage Types (HDD vs. SSD vs. LustreFS) for Different Block Sizes and Different Parallel Read Methods (mmap vs pread vs read)

- ParaGrapher Integrated to LaganLighter

- ParaGrapher Source Code For WebGraph Types